Usually when I am asked to give a few words to describe myself I say macroecologist or large-scale-ecologist. And I might on other days say biodiversity scientist or global change scientist. But a lot of days I would say “ecoinformatician”. Ecoinformatics is the subset of bioinformatics that applies to ecology – that is to say informatic (data) techniques applied to ecology. Some of you may know that I spent 9 years in business before returning to my PhD. But not many know that most of what I was doing was business informatics. Helping companies understand their data. It wasn’t planned. I just have always liked seeing what the data has to tell me. But it turned out to be great training as ecology dived into informatics just as I hit graduate school.
Not surprisingly given my background, I spend a lot of time being asked to make recommendations on how to work with data. I’ve also been involved in some very large data projects like BIEN. Here I don’t want to focus on the large (often social) issues of really big projects (my slides from ESA 2015 on the next 100 years of ecoinformatics are on figshare if you’re interested). Here I want to focus on the much smaller single person or lab-scale project. This post is attempts to summarize what I have learned to be best practices over both my business informatics and ecoinformatics careers. I am intentionally going to stay tool or software agnostic. In this post I really want to emphasize a frame of mind and mental approach that can be implemented in literally dozens of different software packages. In a second post tomorrow, I will give a worked example in R since I know that has the highest popularity in ecology.
Warning – this is a really long post! But I hope it is useful.
I want to emphasize I don’t assume I have the only truth. There are plenty of other knowledgeable people out there (and I hope they chime in). And I’m sure some of them would disagree with me on at least some points. So definitely take what you like and leave the rest. And if this stokes your interest, definitely check out Data Carpentry about arranging classes (but they take no responsibility for what I say in this post as I think they are great but I am not affiliated with them).
So my 10 commandments for managing research data are:
1 – Thou shalt distinguish raw data from analytical data and link them with a repeatable pipeline
One big hurdle I see people stumble on over and over is conceiving of there being one copy of The Data. This may be how the human brain works. But its not how computers work best. Computers are stupid and highly inflexible (albeit very fast). Data needs to be structured for what you want the computer to do with it. I would suggest that at a minimum you should conceive of two types of data (and create both types for a single project):
- Raw data – this is where you do data entry and collect and assemble your data. It will be at the level detail of your measurements and lab notebooks. And it will be structured to minimize data entry errors and promote error checking.
- Analysis data – this is what you feed into various visualization, statistics and analysis routines. This data may be summarized from the raw data (e.g. summarized to weekly or monthly periods). It will almost certainly be organized into a different structure to facilitate analysis rather than data entry. You may even have multiple levels of summary for different analyses (making multiple analysis data sets).
I have seen many people start with raw data and then gradually over time morph it into analysis data without realizing that was what they were doing and in the process destroying the raw data. Much better to have a clean break. The raw data comes straight out of your lab notebooks (or is entered directly into a computer in the field) and then is never modified again. The analysis data is derived through a repeatable pipeline (aka computer scripts that can be run over and over again) that can take the raw data as input and spit out the analysis data as output. This gives you data optimized for the task (entry or analysis) but also gives you essentially one data set from a reproduceability point of view since one button click will regenerate the analysis data.
2 – Thou shalt create raw data in instance-row, variable-column format
Many people like to capture their raw data in crosstab form (Figure 1 – also called spreadsheet style and sometimes table form, but the term “table” is ambiguous so for reasons that will become apparent I am going to call this the dimensional view). But putting raw data in a dimensional view is really confusing raw data and analysis data goals.
I would argue that raw data should ALWAYS be captured in the format where each measurement has one row, and each column is a different variable or attribute (Figure 2 – also confusingly called table view or database or dataframe style) so here called instance-row, variable-column or just row-column format. Variables might be e.g. latitude or longitude where measured or a site code, or a species name or individual code or day collected or abundance or body mass or …
Note that in this case the row-column view and the dimensional view are just different ways of viewing the same data. That is kind of the point of commandment #1. The dimensional view is much more intuitive to analyze (e.g. it quickly makes obvious that Washington has higher abundance than Oregon). It also makes the dimensions obvious (here space and time) as each dimension is one “direction” in the table (horizontal or vertical). But it has many problems for capturing data. First the metadata structure is more obvious in the row-column format (where do the year & site & abundance titles go in the dimensional view). And what if we add species as a 3rd dimension – how do you make that table? And what if we didn’t collect in Oregon 2011? This is obvious as an absent row in the row-column view but how do we show it in the dimensional view? as a 0? an NA? an *? This highlights the power of the row-column view in that every row has an exact 1-1 mapping with a data point. This is not necessarily true in the dimensional view.
Note that there are lots of good places to store row-column format data. Simple CSV (comma separated variable) files, tab-delimited files, R data frames, a SQL table in Postgres or MySQL are all good choices but you can do it in Excel too – its more a mindset than a technology issue.
3 -Thou shalt partially (but no more and no less) normalize raw data into star schema(s)
Data scientists love to talk about normalization. And ecologists are blown away by this concept and do whatever the data scientists say (fully normalize the data). But this is wrong. Most ecology datasets should only be partly normalized.
What is normalization? The concept is easy and might be better phrased in English as factoring. Imagine you are collecting data on individual birds living in lakes (such as sex and body size). You also have some data that only makes sense at the species level (e.g. total abundance of birds observed in the Breeding Bird Survey). And some that is only available at the family level (e.g. diet). You might have a hypotheses tying together sexual dimorphism with abundance and diet. You could store this data in several equivalent ways. Look at the following three figures.
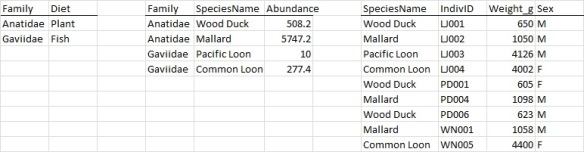
At one extreme we have the top version (Figure 3) which is fully denormalized (i.e. all smashed into one table with lots of redundant information). Ar the other extreme we have the middle version (Figure 4) where the data is nearly fully “normalized” or “factored”* and all redundancies are eliminated by creating separate tables (namely family, species, and observatin tables). Figure 5 is a halfway house and is my recommended approach.
The main advantage of normalization is that it avoids redundant information. Figure 3 repeats the information that a Wood Duck is in the family Anatidae and eats plants 3 times (and also repeats that the average abundance of a Wood Duck is 508.2 three times). Figure 4 only repeats each of these facts once. Note how Figure 4 has a different kind of redundancy – the column Family appears in two tables so the two tables can be linked together (as does species name). From a data entry point of view Figure 4 has a lot to recommend. Often cited is that it is typically smaller. This is a worthless reason – disk space is cheap and most ecological datasets are quite small. But a much more valid argument is that fewer redundancies are fewer opportunities for errors in data entry to creep in. But in my experience computers may be great at working with denormalized data but humans get lost quickly as the number of tables grow. So, I find that as is so often the case, compromise is the best option. So I favor partial denormalized data (Figure 5, the last example).
In fact I favor I very specific schema (that is the database techie word for the design of data tables to use) that have been around in the business world for decades known as a “star schema“, which is partially de-normalized. A star schema has one central row-column table known as the “fact” table. In this table each row corresponds to measurement on one entity. And the columns of the “fact table” are either dimensions (to be defined in a minute) or measurements (like body mass and sex). The right-most table in Figure 5 (or Figure 4) is a fact table. The “dimensions” are the “coordinates” of the facts. In ecology space, time and taxa are extremely common dimensions. Figures 3-5 have only one dimension (taxa). Figures 1 and 2 have two dimensions (space, time).
A star schema therefore has one fact table and one table for each dimension. The fact tables are linked to the dimension tables by unique identifiers like species name, date, site code, or site lat/lon (i.e. these columns are duplicated and found in both the fact table and one dimension table). Note that a dimension table will have many fewer rows than the fact table. Indeed the space dimension table will have one entry for each site that you observed at. The taxa dimension will have one entry for each taxa (or individual) that you observed. Thus if you have say 20 sites, 100 species, and 3 years you will have one space dimension table with 20 rows, one taxa dimension table with 100 rows, and one time dimension table with 3 rows. Your fact table could have as many as 20*100*3=6,000 rows (if every species is found at every site every year), but in all likelihood will have somewhat fewer rows. To me this is the “right” amount of denormalization or factoring. Basic attributes of a species are “normalized” and pulled out in the taxa dimension table and not repeated for every observation. But families and species are denormalized (aka repeated) in the fairly small dimension tables leading to a little bit of redundancy. Again this is called a “star schema”. An example is in Figure 6.
In this figure each rectangle is one row-column table. The column names are listed in the rectangle. In this example “Observation” is the fact table. It stores one measurement, abundance, and three dimensions (taxa/Species, time/DateObserved, and space/Site). Note that the Site dimension table is linked to the fact table by the shared column “SiteID”. This is denoted in this type of drawing (again known as a database schema) by the line linking SiteID in the two tables. The “crowsfeet” shows the direction of the many-to-one relation (i.e. there are many entries in the Observation table for one entry in the Site table). If you want you can use software known as Entity-Relationship or ER software to draw these diagrams. I used the free program ER Assistant to draw this. But that is probably overkill for most people – pen and paper is more efficient unless you do this a lot.
The right way to use a star schema in data entry is the followin:
- Figure out the dimensions of your data (space, time and taxa are usual suspects but you may have more or less)
- Create your dimension tables (the small tables that surround your fact table) first. Some people would call this a data dictionary or in social sciences “the codebook”.
- Enter your data into the fact table
- Either in real time as you enter the data, or periodically, check your entries in the fact table to make sure they actually point to entries in the dimension tables (this can be done by join operations which are discussed below).
The origins of the name “star schema” may be obvious in Figure 6 – it has one big central table and lots of little tables coming off it. It looks (if you squint) like a star. And in case you are wondering, you may have more than one fact table. A common example is where you have some organismal measurements (abundance, body size) with space, time and taxa dimensions. And then you have environmental measurements (temperature, soil pH) with space and time dimensions. It might not surprise you given how cute computer scientists think they are that a star schema with multiple fact tables is called a “constellation schema”. My post tomorrow giving an example in R on this topic will use a constellation schema (one biotic fact table, one environmental fact table). If you’re into cute names, if you take a star schema but then fully factor/normalize the dimension tables into multiple tables (go from Figure 5 to Figure 4) it is called a snowflake schema (you have to really squint to see a snowflake) (But don’t let computer scientists talk you into using snowflake schemas!).

Figure 7 – Snowflake schema with 3 dimensions (space, time, taxa) but the dimension tables are normalized
Note that again a star schema can be implemented anywhere that can implement row-column data. For example, each fact or dimension table can be a separate CSV file. Or they can be separate tables in a SQL database. Or separate tabs in an Excel spreadsheet. Again, its a mindset, not a technology.
4 – Thou shalt plan ahead to spend a lot of time cleaning your data
Over twenty years ago, Gartner group (a consulting company that advises business on how to keep up with computer technology) said that 70% of the work in building a data warehouse was cleaning the data. Now datawarehouse is the business buzz word for what I called analysis data above. Businesses capture raw data in transactional systems like cash register point of sale systems, accounting software, etc. This then gets transformed (by a reproducible pipeline) into what business calls a datawarehouse and what I call analysis data. Gartner’s point is you might think if you already have the data in one form it should be quick to go to the other. But Gartner estimated that 70% of the work (i.e. a huge amount of work) goes into cleaning up that transactional data. Store A gives product X one barcode, while Store B gives the same product X a different barcode. And these have to be reconciled. Or field assistant A calls a species by its old name, and field assistant B uses the newest taxonomic revision. Or field assistant A enters states using their two letter postal codes (Oregon is OR), but field assistant B is from Europe and makes up 2 letter codes, or is American and forgets whether MI is Michigan or Mississippi or spells them all out. But in the end, long story short, expect to spend at least half your project from data entry to anlaysis cleaning up your data. Plan for it. Do it up front. Don’t just fix things when they give you funny results because then you won’t have fixed a lot of things that give you non-obviously wrong results.
As an aside, note that one of the benefits of treating raw data separate from analysis data is you can do somethings to minimize these problems. As discussed above, you can easily write code that checks that every entry (row) in the fact table matches out to an entry in each dimension table (e.g. if I enter a fact with state as “Miss” is there a row in the space dimension table that has a state of “Miss” (probably not since this is a non-standard abbreviation). If you’re really fancy you can do these checks real time as somebody enters the data. Note that this assumes that you create your dimension tables first, then your fact table.
5 – Thou shalt not hand-edit raw data
Once entered – you should never change your raw data again. All those errors you find in step #4 (e.g. entering “Miss” instead of “MS” for the state of Mississippi) should be modified and corrected in a new “corrected raw data” copy of the data populated by scripts. It might seem clunky to go find all the entries of MI in state for field assistant B and change them to MS instead of just go edit them by hand. But you will regret it if you edit by hand in the long run. Two main benefits of putting it in a script: 1) All those times you thought you found an error and “fix” them only to discover you misunderstood and introduced an error are safely covered because you still have the raw data. 2) You have documented everything you have done and the corrections are reproducible (and reusable if a new year of raw data comes in). This inherently requires a programming language, but there are still many choices. There are tons of tools for data cleaning. In business they go under the generic name of ETL (extract, transform and load). Data Carpentry uses OpenRefine. I like awk. Many bioinformaticians prefer PERL. You could even write VBA scripts in Excel. And tomorrow I will give some examples in R. But hand editing Excel spreadsheets is right out. It has to be a script you can run over and over.
6 – Thou shalt conceptualize analyses as dimensional
What shape is your data? This may seem like an odd question (most would answer “rectangular” if they had any answer at all). But when you start thinking dimensionally it becomes a more relevant question. When thinking about analyzing data, I find it very helpful to think about it as a multidimensional cube:
In Figure 8 – the fact table is visualized as a cube with three dimensions of time, space/site and species. Each “cell” in this cube holds an abundance for the particular time/site/species combination it is at the intersection of. This makes the notion of dimensions as the “coordinates” of the data very explicit. And the dimension tables are shown as ancillary tables (time doesn’t have one here and species has a phylogeny overlayed as well).
Remember when I explained why the dimensional (crosstab/spreadhseet) view of data had problems when you go into three dimensions in Commandment #2? Well it does have problems when you try to represent in say Excel. But you can visualize and conceptualize it really well. And in fact this is how I conceptualize every dataset I work with. It then becomes really obvious to start thinking how to summarize data. Perhaps you want to average across time, and sum across sites, leaving a vector (1-dimensional list) of species (which can still be easily aligned with the taxa dimension table). And so on. You can still do all of these operations on a row-column table (and per commandment #7 below you may end up doing exactly this). But I find it easier to think about data dimensionally. Of course I might be biased since I spent several years as the product manager of a multidimensional database named Acumate (now defunct but a detailed description of multidimensional databases can be found here albeit in a business context). For a while there was also a market for multidmensional spreadsheets.
Just like you can have multiple fact tables you can have different multidimensional cubes (and they may not all have the same number of dimensions). This is a good way to think about ways in which your different fact tables overlap and don’t overlap. Here is a two fact table/cube representation of the constellation schema I showed before (Figure 7).
7 – Thou shalt store analytical data as denormalized but optimal for the tool you are using
Analysis should ALWAYS be done on a fully denormalized data (i.e.like figure 3) where everything is mashed into one big table and redundant. Your star schema that you used for data entry should be collapsed back into a single data table for analysis. You do this by a process known in the database world as a “join“. A join is simply combining two tables by a field they have in common (e.g. SpeciesName in the example above). The join is a horizontal concatenation – i.e the tables are joined side by side or to put it in other terms – every column in table 1 and every column in table 2 become columns in the joined table (except the joining column which is not duplicated but shows up only once in the joined column). It is basically going from Figure 5 to Figure 3 (i.e. a a join is the opposite of a normalization or factorization). In a star schema you will still have the same number of rows as your fact table, you will just have a lot more columns in your single denormalized table. In theory one can do joins on the fly depending on the analysis you want. But I say why bother. Once you have your data cleaned up in a star schema, do one last step and join it all together into one big table.
You can do joins in any software. You can use the merge command in R. Or you will see in the next post I recommend using dplyr and join commands in R. You can do joins in SQL (the syntax is beyond this post but found in any introduction to SQL). You can do joins in Excel using lookup tables. Its a core concept in data management.
How should you store your one big table for analysis? Some software directly supports the multidimensional view. For example n-D arrays in Matlab or NumPy arrays in Python. In theory you could do it with n-D matrices in R but R doesn’t really like n-D matrices and one of the problems with cubes is they explode very quickly (100 species, 100 sites, 100 times gives 1,000,000 cells) and R’s poor memory management bogs down quickly. In other software you are much better to keep the data actually in row-column form but just think of it multidimensionally. Dataframes in R or Pandas data in Python are good examples of this. I actually use a mix of row-column and multidimensional cube storage in Matlab when I am coding. Usually I start with row-column when the data is really big and after I’ve filtered it I convert it into a muldimensional cube, then summarize it and run statistics on it. The point is you can have a conceptual model that is multidimensional and a data model that makes your analysis software happy.
You can do dimensional-like summarization in any software. They’re called pivot tables in Excel. They are the apply family of commands or the by command in R combined with commands like sum and mean. Or in dplyr package in R they are group_by and summarise commands. These have direct analogies to the group by and summary commands in SQL.
8 – Thou shalt create analytical data and analyze it by reproducible code
If there has been one consistent theme, its don’t ever do anything as a one off. Yes you can create an Excel pivot table from a star schema and summarize it using some points and clicks on menus and dialogue boxes and save the results out. But can you reproduce it if you get new data? Everything to get to the fully denormalized analysis table should be done by scripts that can be run repeatedly and reproducibly and reusably.
9 – Though shalt obsessively hand check the transformation from raw data to analytical data
So you may have noticed that I recommend a number of steps to get from raw data entry to analysis. Thou shalt never assume that simply because the computer code ran and spit out data of the right shape you have the right answer. You almost certainly don’t. There are all kinds of ways things can go wrong. It is critically important, indeed, I would say a matter of scientific ethics, that you laboriously hand check your data. Take a row in your analysis table and hand trace it back to the raw data. Can you reproduce that row from the raw data by hand calculations? If so take another entry and trace it back. Do this enough to be really confident it is right. Similarly scrutinize your analysis table. Do you have a species called “NA” (or a time period or site)? How did that sneak in there? Is an endangered species showing up as your most common species? Did your joins work so that a species is lined up with the right family? Really poke and prod your data. Torture your data to make it confess its errors (a variation of a Ronald Coase quote)
10 – Thou shalt use the simplest feasible technologies to keep the focus on ecology
You will I am sure have noticed that I have repeatedly emphasized that the important thing is the mindset and approach not the technology. You can do everything I suggest here in Excel. You can also do everything I suggest here in an SQL database. Which should you use? Well you have to weigh it out. In particular I think the main trade-off is learning-curve vs scaling. Using more complicated technologies (like an SQL database) is a steep learning curve for almost all ecologists. This weighs heavily towards using familiar tools. But the bigger and more complicated your data is, the more that learning curve starts to pay off in the ease with which these concepts can be put into practice (and the automatic error checking built into more advanced tools). In short, the bigger and more complicated your data, the more it is likely it is worth the learning curve of a newer more sophisticated tool. But don’t let a computer savvy person intimidate you into saying you have to do data management in an SQL database. Or that you have to normalize your data.
Summary
So overall I have been arguing for a workflow (or pipeline if you prefer) that looks like (Figure 10):
So you will start with some dimension tables (probably created by hand in advance) and one fact table (“Raw data in Star Schema”) that will contain errors. You will clean these errors up by scripts which will create a new star schema that is almost identical in size and shape (“Cleaned Data in Star Schema”) and then you will denormalize it and end up with one mega row-column table or multidimensional cube (“Denormalized Analysis Data”). Note that this pipeline calls for 3 “copies” of the data. I put copies in quotes because they are certainly copies (i.e. distinct files that contain redundant information) but if you follow the text on the arrow and create these copies not by hand but by scripts that can be rerun, then in many ways these are really just 3 different views on the data, not 3 copies.
Tomorrow I will post an example dataset and an example R script that moves through the data in agreement with these 10 commandments.
I’m curious. For those of you who think a lot about ecoinformatics, what do you agree or disagree with? And for those of you who are new to these concepts, what have I explained poorly? What needs expansion?
UPDATE – a direct link to the next day’s post with sample R code
UPDATE 2 – Eric Lind pointed out this nice paper by Elizabeth Borer et al on data management which matches many of the themes herein.
* I say nearly fully normalized because a real hard core normalizer would recognize in the real world we would probably have 60 families and only 5-10 diet types and recommend that diet-type be its own table and family would then have codes pointing into the diet type table. That way there is a master table of diet types where each diet is entered once and spelling scan be checked against it.









Brian,
I’ve never seen this laid out so explicitly for ecologists. It will be fun to use in our weekly grad workshop for first semester students, Advanced EEB.
My only comment is that field collections add a complementary activity–the curation of specimens/soil samples/root cores/photographs. In short, analog data. The same principles apply: never throw out a label, even if you reprint it. Keep old and new labels together with the specimen. If you process a photo, keep the raw image. Maintain and deposit voucher specimens. Separate raw data (specimen, or at least it’s tissue) from analyzed data (DNA barcodes, elemental chemistry, stable isotopes).
And just as the media of digital data (remember tape drives, punch cards, floppy disks, Zip drives…?) need to be curated, biological specimens need to be stored, the ethanol topped off, the pinned specimens checked for dermistids. This is a lesson I continue to learn, sadly. A life in field biology generates a lab that can increasingly resemble your garage or attic, if you are not careful. I think we have 9 freezers and refrigerators full of berlese samples, soil samples, in thousands of nested plastic bags and vials.
The extra workload of storing raw specimens (warehousing), and curating analog data was a big point of contention in the evolution of NEON, whose DNA evolved over time from biological survey, to digital maps of abiotic measures, to the hybrid we see today. One of the big tasks ahead comes in generating protocols/workflows on how to allocate and work with soil samples, pitfall trap catches, etc. Entomological types are familiar with this problem as “working with the by catch”.
But as I said at the beginning, much of it rests on Brian’s 10 commandments, However, you left out 11: You’re all individuals! You’re all different!
Thanks Mike,
Very interesting but now that you say it totally intuitive points about the parallels with “analog” data. And I’d never heard it but it makes sense about the issue with NEON.
I fully endorse commandment 11 as well.
Such an important issue! I look forward to seeing your worked example later.
After a lot of time hanging around with historians and archeologists, I’ve integrated the museological concept of provenance to my data management. So every set of observations is also annotated with a provenance field that states where the data come from – whether this is extracted from a publication, a particular instrumental analysis, or from another person, or a specific model. The provenance fields can be as simple as a pointer to a publication or longish descriptions that reconstruct the history of a source. This type of information is so important for parameterizing models and evaluating bias, establishing limits of detection, observation reproducibility, and errors when working with heterogeneous data sets.
I agree that provenance is a very common and important thing to track in many types of data. It is, ultimately, part of what it means to be a scholar.
Worth thinking about molten data for storing data, which is very flexible, though requires some thought to “cast”: https://www.r-bloggers.com/melt/
melt is another good choice in R. I have used it, but it is the one I’ve used least personally. As somebody who knew SQL before R, and appreciating the cleanness of design to the Wickham universe, I find dplyr/tidyr more intuitive for myself. But its totally a case of personal preference (and I suspect what one learns first). People should definitely check out melt. Thanks for the suggestion!
Excellent and well-thought/ presented topic, Brian! I don’t know as you would allow for introduction of commandment subsections- but I offer one up for consideration. 4(a): Thou shalt invest considerable time in the “cleaning of” field personnel. I learned this the hard way, when I had several field crews collecting data at the beginning of a multi-year monitoring project. Sheesh- what a mess, as it took nearly 3 months to clean-up what they had entered into our field data forms. There were relatively few “human errors” on these data sheets. The problems were rooted in some of the things you mentioned. Different people calling species X by a variety of scientific/ common names; unidentified species being vouchered in a variety of ways; site identification codes having unique identifiers- and so on and so forth. It was a nightmare getting it all straightened out, really.
Since then, I have spent MUCH more time including field techs in the planning stages of a project (especially the decision-making process for data acquisition & entry). I also have spent MUCH more time in the field, especially at the beginning of projects, with the field techs. And, I always make time for a “dry run”- where I will have field crews gather data for a day or two (yes, it is an added expense). I also train and appoint a “data-monger” for each field crew. This person reviews each and every data form on the evening of the day it was collected, and performs other tasks (such as photographing and sending me data as it is acquired)) so I can review it in “real-time.” Combined, this approach probably eliminates upwards of 95% of the mess that would otherwise require an after-the-fact and clumsy “clean-up.”
Great tips! There is definitely a theme here that data curation needs to be an effortful, thoughtful process from start to finish.
Great post Brian, thanks for sharing and looking forward to the sequel. (Also appreciated your ESA slides, though sad to have missed the narration). You cover lots of important issues that are too rarely discussed, and worse, issues that are too often ignored as trivial or irrelevant.
I agree wholeheartedly with almost all of your commandments, though I might wish you had touched on a few other things (metadata, provenance, data versions, which you may have covered in your talk — maybe fodder for another awesome post?)
I did want to query your further on non-normalization and star schema. At this level, why not just ignore the star schema piece and gun for one big table? Like you, I agree that analyzing ‘one big table’ is almost always preferable in the ecological context, rather than insisting all joins be done on the fly. So why bother with multiple tables normalized to any degree? We discuss this issue in rOpenSci occasionally regarding the best return formats, and the consensus has generally been that users almost always prefer a pre-joined table. After all, the data is rarely so big that this approach is impractical (if it were, analysis step would likewise need to do joins on the fly), and we find many users dislike or are unpracticed doing their own joins all the time. If the main reason for some normalization is to save space (and only on disk at that) it seems at first pass we can just go for one big table, yes?
On the other hand, some data is so nested that a single table really doesn’t make sense. This is particularly common when data comes from some nested list format (e.g. lists & other R objects, json, xml). I’ve been interested to see Hadley’s recent directions in making heavy use of data.frames whose cells are themselves complex types (e.g. https://blog.rstudio.org/2015/09/29/purrr-0-1-0/) which feels wrong to me (how would you store such data.frame as a csv?) but also offers some very elegant abilities for analysis that otherwise gets very cumbersome.
Also I do worry that as ecoinformaticists we sometimes give the impression that it is always intuitive what ‘column variable, row observation’ means. If I fit a model to a series of datasets, I might make a row for each dataset, and a column for the corresponding parameters; e.g. columns like: ‘dataset’, ‘par 1’, ‘par 2’. If I then repeat this, fitting a different model with different parameters, all of a sudden it might make more sense to use a ‘longer’ form, where columns are ‘dataset’, ‘model’, ‘parameter name’, ‘value’ (i.e. tidyr::gather / reshape2::melt), than the natural join of the two tables (i.e. columns of dataset, modelA_par1, modelB_par1, ..).
Oh, one other thing — I wish one of the commandments was about dates. How do you feel about dates being stored across separate columns (e.g. year, month, day), instead of one ‘date’? Seems very common in ecological data and has always bothered me.
Hi Carl – thanks for all the substantive comments. I know you’ve thought deeply about these issues yourself. My thoughts:
1) Why partially normalize (i.e. star schema). For me this really comes down to the avoidance of redundancy on data entry and the ability to do error checking on things like site code or species name. One is effectively requiring the creation of a data dictionary for these types of things. Some people would even call this metadata (although I wouldn’t). I think the discipline of creating a data-dictionary/dimension table requires one to think through what are acceptable values. Which can then be used for error checking (real time or post hoc) in data entry. If you just enter a denormalized data table up front not only are you redundantly reentering data on say species attributes like abundance or family, you are certainly going to end up with a list of species names only some of which are valid and you have not yet declared anywhere which ones are valid or not.
I fully agree with you though that the average scientist/ecologist does not like joins and just wants the end goal which is always denormalized. I guess I would say if you still have data entry/error checking the star schema is worth the trouble. If your error checking is already done (which I think may be the scenario you describe for your rOpenSci return values) there is no real reason to use the star schema and just go to denormalized. i agree space savings is an often given but practically unimportant reason.
2) Matlab is really fond of the structure within a structure paradigm as well (they have cell arrays which are a bit like lists in R). I use them occasionally, but honestly I cannot imagine very many ecologists getting their head around them. Also, I just think the dataframe/table with an extra column (like your example of dataset) is not only more intuitive but more flexible (it lets you change what is your innermost nesting – maybe you want to run a regression on each dataset, but maybe you want to run a regression across datasets within a state – easy to do in flat table but not in nested substructures).
3) Your examples of what should be a column are good ones. And as I’m sure you know they fall under discussions of degree of normalization in databases (e.g. 3rd normal vs 5th normal). I don’t have a good answer except to say that I think ecologists will figure this out faster if we: a) don’t normalize too much, b) keep giving them examples, and c) encourage them to actually draw out a schema with all of their variables before starting to collect data – something that is rarely done but I think not unreasonable to do (any more than dissertation committees want to know which statistical analyses will be used). In my experience most ecologists have a much easier time with parameter1, parameter2 columns than parameterrank and value columns. But there is more flexibility in the latter form. I guess it goes back to trade-offs of learning curve vs flexibility.
4) I’m totally with you on dates – storing them as separate columns makes little sense to me – especially as you can quickly recover month of year if you need (as show in my example tomorrow). Store it in a single column in an unambiguous format 13Mar2016 or 2016Mar13 being one format that will be read the same by Europeans and Americans. Then any good software (such as lubridate in R) can quickly convert this to an internal format that the software can do date arithmetic on.
Great piece, applicable across so many domaines!
Brief note on dates — after working 20 years in the US, four in Sweden and a bit in France I endorse “YYYY-mm-dd” over using a letter abbreviation of month. If a date begins with a four-digit year it’s ALWAYS followed by month, and you don’t risk spelling confusion. Obviously France and the US are at odds when you begin with either month (US) or day (Euro-ish-style), but beginning with four-digit year makes the format explicit.
Additionally, this matches an explicit date type in any database, which can speed up queries done within the database.
Interesting point about dates. I guess it depends on the goal. As you say YYYY-mm-dd is a “native” style to most computers and avoids language confusion on month. Perhaps to a non-techie it is not obvious that it is YYYY-mm-dd instead YYYY-dd-mm (I know that is a nonsensical format never seen but its not obvious without some thinking).the advantage of 2016-May-13 is no human will ever confuse what it is (even if they don’t speak the language) so it is a bit of metadata. I guess its a trade-off depending on goals, but I suspect you’re right that more often the YYYY-mm-dd is probably the best. Thanks for stopping in!
Glad to see someone bring up dates! Excel’s tendency to change date formats when re-saving CSVs is likely the largest single source of errors in my office. I usually use YYYY-mm-dd format (like in the comment below) but I’d never considered the month name option. I’ll have to ask my coworkers what they think.
“Note that there are lots of good places to store row-column format data. Simple CSV (comma separated variable) files, tab-delimited files, R data frames, a SQL table in Postgres or MySQL are all good choices but you can do it in Excel too – its more a mindset than a technology issue.”
Eh, except when you happen to pick a technology that is not easily accessible because of e.g. an obscure, proprietary format that may not be accessible in the future. Stick to openly documented, easily readable formats — or at least have a backup in such a format.
In theory I agree and this is important. In practice, the only proprietary format in my list is Excel which I expect is what you’re objecting to, and honestly Excel is not going to go away anytime soon. I can still open Excel 1997 and even dbf and Paradox (if I want to really date myself) files with a few clicks with tools on almost everybody’s laptop.
Given how many people double click a CSV file and have it open in Excel automatically which can then corrupt the CSV if they accidentally hit save (e.g. changing quoting structure or dates – see this post http://ecologybits.com/index.php/2016/07/06/beware-this-scary-thing-excel-can-do-to-your-data/), there are risks to every file format.
I’d be careful with “Excel is not going to go away anytime soon” — it is a matter of temporal horizon you have in mind. Who knows what happens in 50 years, and what kinds of revolutions are ahead of us. We are already almost unable to retrieve stuff from 3.5 or 5.25 floppy disks. Or a more radical example: it was a sheer luck that someone has left us the Rosetta Stone (hard, durable stone), without it we’d be unable to open the data of ancient Egyptians. In 500 years there may be no stone to decipher the bits of our data that used to be stored Amazon cloud and stored in a bizzare xml-based format called .xlsx.
With long-term view on data usability, I think that the least we can do for our kids is to store the data in formats that are as simple and open as possible. Since .csv is simpler than .xlsx (or .dbf), and it is not proprietary, .csv is more sustainable and should perhaps be preferred. At least for data publications and supplementary materials.
As for the data structure (normalized vs. denormalized), maybe we should also think about which of the two structures better resists the dent of time. In 500 years, when post-apocalyptic people are scraping fragments of data from harddrives in ancient underground bunkers, what is the format that can be more easily restored, the one with redundancy, or without?
@Peter Keil:
I’m going to take what might be a contrarian position and say that we shouldn’t bother trying to anticipate the needs and technical capabilities of distant generations.
I mean, in what concrete ways do you feel worse off because you can’t access the data that past ecologists stored in a now-unreadable format on 5.25″ inch floppies? Do you ever have occasion to curse their lack of foresight? And if you did, wouldn’t that be awfully unfair of you?
It’s *very* unlikely that *anyone* a century or two from now will want *any* bit of data you might collect. After all, it’s highly unlikely that anyone *today* (besides you) will want any bit of data you might collect! Any “what format is your data stored in” is, I suspect, far from the only or even most important determinant of whether an ecologist in the distant future will access your data.
The “needs” and “wants” of long-distant future people aren’t well-defined today. What they “need”, and “want”, and what they do to meet those needs and wants, will depend on what we do today–but in complex and unpredictable ways. At the timescales you’re talking about, there’s nothing but Rosetta Stones. Nothing but sheer luck.
I’ve got nothing against CSV. I prefer it over Excel myself and generally recommend it. But I also can’t really get on board with the Excel is evil bandwagon either. The only time horizon I care about/think is realistic is 5-10 years. Beyond that changes in physical storage (as you noted and I was tempted to note in my reply to SomeGuy), changes in data measurement standards, retirements followed by desk purges, etc, are all bigger. My point is that over a 5-10 or even 20 year horizon, Excel has been extraordinarily durable. There is a certain lock-in. Even though Google wants to annihilate Microsoft and Excel, their spreadsheet program reads Excel. And its successor will too. And Excel has more metadata potential than a CSV file.
To repeat myself, I prefer and recommend CSV too. But I can’t agree that Excel is a bad choice.
Just to illustrate why Jeremy’s contrarian position is (IMHO) wrong, I recently needed the raw count data of a classic sub-fossil pollen dataset to test/illustrate how a machine learning method I’ve been looking into with a post-doc works. Using a well-studied classic data set is a good way to compare methods and check what new methods are doing. Most people used this classic pollen data set in percentage format (normalized by row totals) and that is the format I have always had a copy of the data in. This new method needed the raw counts. I was sent the data by the originator but this data set, being from the 1970s (collected before I was born, just) was in some obscure format that could only be read by a very old DOS-based executable that I doubt would compile easily compile on a modern machine even if I had the source code (which I don’t). So, now I’m writing my own R function to parse the raw count file (thank some Deity it was stored as ASCII text not in some binary format!), which seems like an epic waste of my time.
Just to be clear – I don’t blame the originator for storing the data this way. There were real constraints on data storage when this file format was used. Today we don’t have such issues (for most ecological problems anyway; distributing the few massive data sets aside) and there is no justifiable reason for doing so, beyond the personal time/effort one.
As for hardware; I am aware of several instances where data has had to be recreated from original records or was lost entirely because of 5.25″ discs. I don’t think I now have access to a machine with a 3.5″ floppy drive either. Yes, I’m sure even now I could find either of those drives and recover data from this kind of media, but at what cost in time, effort, or money? The problem is that these discs sit at the bottom of a desk or filing cabinet drawer for years or decades, they might even be thrown out during a move. They are generally forgotten, right up to the point where someone really does need the data.
I’m sure we all have personal experience of problems of this sort, or know someone who has had them. We have the technology and the standards now to avoid repeating these problems in the future. The one point I think Jeremy nails is that we don’t know what future researchers or societies will want or need. But where his contrarian’s view would be to throw up one’s hands and not bother with securing data in open formats, we should proceed from the point of view of assuming that someone might want the data in the future and that therefore we should do our best to preserve those data in open formats, archived in proper digital repositories.
I don’t have anything substantive to say, as this is all totally news to me. Which is actually my point: whenever Brian or Meg blogs about something I’m totally ignorant about, it’s the most pleasant possible reminder of my own narrow horizons. I mean, Brian and I share a lot of interests, have had many similar experiences–and yet here’s something he considers totally basic and widely familiar that is *completely* novel to me.
(Aside: I confess I consider myself fortunate not to have to worry about most of the stuff the post discusses. A side benefit of being an experimentalist who often works solo is that my data all fit very naturally into one small spreadsheet from the get-go. In my lab, we literally just transcribe our data sheets into Excel.)
Thanks Brian for the clear post with very good advice! A perhaps important commandment that I missed is related to data preservation. The amount of data (both published and unpublished) that are lost forever is simply horrifying. Vines et al (http://dx.doi.org/10.1016/j.cub.2013.11.014) showed that data availability for published studies decreases with time at an alarming rate of 17% per year. Too much data rest in a single computer or hard drive and are lost with hardware failures or changes (who can recover data from a floppy disk nowadays?). I know very few ecologists (if any) who haven’t lost data in one way or another (myself included).
So I’d stress the importance of storing data appropriately from the very beginning, backed up in appropriate formats and probably in the cloud (GitHub, Dropbox, Figshare, etc). Hart et al, for example, compile advice on this (https://doi.org/10.7287/peerj.preprints.1448v2).
Good point and useful tips.
Gavin, Jeremy & Petr
Important issues about data availability (which I took as beyond the scope of my post, but there is no reason you have to).
Gavin – I agree with you on this. I have definitely asked people to dig out datasets over a decade old, and to productive effect as well. It definitely happens. And science would be better off it could happen more. I’m not sure if I can agree with Petr’s image (somewhat tongue-in-cheek I am sure) of random people 100 years from now. But people should plan to make sure their data will live for at least a couple of decades
It seems to me the three barriers are:
1) Physical media – yup I have files from college on a Mac 3.5″ disc I would love to recover. This is a big one. The “cloud” takes care of it as long as the funding for whatever piece of the cloud you use exists. To me only time will tell if this is a longer or shorter life cycle than a magnetic storage medium format.
2) File format – your story sounds horrendous. But I’m not sold that really widely disseminated proprietary formats are inherently much worse than “open” formats (especially when the alternative is not bothering to publish at all). But I lean to open formats.
3) Understanding the data – this is a function of forgetting then retiring from the original researcher. The main answer here is extensive metadata. This to me I think is an enforcement issue. Ecological Archives enforces strong metadata. Few other places have the time/resources/ability to do that and the result is very mixed in my experience.
I guess to me if I listed what I’m most worried about in order it would be #3 biggest worry, then #1, then #2, so I’m always surprised when people focus on #2. But just my 2 cents.
fwiw, I always recommend storing raw data as .csv. It’s common sense that pure text files are best for long-term compatibility, and I don’t see the downside – easier to handle with R, can click-open with OO / LibreOffice, and if you must use Excel for viewing just make a xlsx copy (or maybe you have one already, because you typed in the data as xlsx).
I agree the file format is likely not the most important issue for keeping the data alive, but I would say #1 is basically solved (permanent archives, enforced by journals) and #3 is difficult. #2 is a long-hanging fruit, so why not pick it?
Re: #1 being solved, depending on what you consider “solved”, #1 was solved long ago: we can still read centuries-old books printed on paper, as long as they didn’t get destroyed in a flood or fire or whatever. 🙂
Forgot to say that I liked the post, really good advice!
Hi Brian, great post again. I am coming a bit late, but here it is with emphasis on R.
I fully agree with #4 and even blocking out time for data cleaning/processing turns out to be an underestimate (i.e. assuming 1 week becomes 2 weeks or more due to digging deeper and deeper to track down issues). This is especially cumbersome in the biodiversity monitoring context where data flows in continuously and results are expected based on ‘best available info’ to be used in resource management (see recent papers here and here). Some says this is the surest way of making ourselves indispensable 🙂
My workflow is just like you described in Fig. 10 (raw –> star –> denormalized) and I prefer using packages/functions that do one thing very effectively (these happen to be mostly my packages/functions that I wrote to solve the very issues at hand). Thus I tend to avoid general purpose packages/functions (like aggregate, by, reshape, dplyr). For example my mefa4 package is extremely fast at making sparse pivot tables (Xtab function: I use it with several 10^5-10^6 data points) and grouping data in pivot tables (groupSums and groupMeans functions: I use them with 10^6 x 3600 tables). The point here is that sum and mean is what I need most of the time, and it makes a difference to use specialized code (I did some performance comparison). Here and here are papers describing handling data dimensionality in this context if you are interested.
Finally, I am in the process of creating a fake data set for an R workshop that is stuffed full of all the conceivable errors one can imagine. People have mentioned Excel garbling up things, or issues with dates. I’d like to hear more, here is the link for comments: http://peter.solymos.org/etc/2016/06/14/data-set-with-all-the-conceivable-errors.html
Great to know about the mefa4 package.
I’m going to give you credit for slipping a typo/misspelling into commandment 9, which is about obsessively checking for missteps. Well played. 🙂
I’ll take credit whether deserved or not!
Pingback: How [and why] should early career researchers engage with open science? – Callum J. Macgregor
Pingback: Utilizing the R package ‘swirl’ to learn R’s ‘dpylr’, ‘tidyr’ and ‘lubridate’. | Ecological Relationships
Excel was mentioned quite a bit in this post, here is a recent paper on the topic:
Gene name errors are widespread in the scientific literature
http://genomebiology.biomedcentral.com/articles/10.1186/s13059-016-1044-7
https://www.washingtonpost.com/news/wonk/wp/2016/08/26/an-alarming-number-of-scientific-papers-contain-excel-errors/
Pingback: Por que ler blogs? – Mais Um Blog de Ecologia e Estatística
Pingback: » Will future generations be able to read our e-papers and use our e-data?
Great points and useful tips!
Pingback: Book review: How the Hippies Saved Physics | Dynamic Ecology
Thanks Brian for this awesome and complete post. I would add that some raw data type should require extra pieces of information. For instance, plot coordinates should never be stored without a projection system (we have to use EPSG or SRID keys). Moreover, store Taxonomic Serial Number into species table is also a good practice to track future change in species taxonomy.
Great blog!
“Dimension” is used in a way that is unfamiliar to me. I wonder whether it corresponds to what I think of as operational factors in analytical laboratory work, e.g., facility, operator, assay run, etc. If so, when modeling reported values, we treat those factors as random effects and on that basis estimate method precision components (repeatability, reproducibility, and intermediate precision).
Say, for example, we have constructed a series of test samples by dilution for the purpose of describing the precision components. We are going to fit a model of the concentration- (or dose-) response relationship. The data would be observations of:
• y = reported outcome, binary (logistic regression) or continuous (4-parameter logistic function)
with fixed effect
• x = concentration (continuous, log-transformed)
and other attributes
• facility: Campuses A, B, C, …
• operators A1, A2, A3, …, B1, …, C3, … (operator nested in facility)
• runs A1-Monday, A1-Tuesday, …, C3-Wednesday, … (run nested in operator)
Looking downstream to the analysis and interpretation of the mixed effects model,
• repeatability (intra-assay precision) is residual variance
• intermediate precision includes also variance attributable to run and operator and
• reproducibility also includes variance attributable to facility.
What do you think? If that seems right, then 1) I can hitch my understanding of the commandment to “conceptualize analyses as dimensional” to that system, which is familiar to me and 2) the correspondence between dimensions and random effects may be a helpful insight for other readers.
Of course dimensions (e.g., site and date in your hawk abundance analysis, or summaries of them like region and month-of-the-year) may, for some purposes, be treated as fixed effects, so I see it is not a perfect correspondence. But can I at least say that factors I anticipate including in my model as random effects will be “dimensions?”
Yes dimension is a rather abstract term. I guess I would say they are aspects of the measurement but not the measurement. So yes things like lab, operator, and run would all be examples. They do often have aspects of space or time or types (species, products). There is no reason to pull them out as a separate dimension though unless you have data on those entities. So for example if you have data about each lab (model of machine) or operator (e.g. years of experience) then it would be useful to pull it out into a separate table/data file. Otherwise, if you just keeping tack of the lab and nothing more about that lab, then you are probably in the scenario Jeremy described in his comment, and you may just keep one big master table and not pull out a separate lab table/file (although just to contradict myself having a separate table that has a master list of labs can be a useful way of crosschecking if there are misspellings of lab IDs in the big master file).
Thank you for your reply. I appreciate your points about how rich the information is about the dimensions.
I hate to nag, but what do you think about the connection between dimensions and (anticipated) random effects? (I put “anticipated” in there because I’m cognizant that we’re just talking about preparing data for analysis, not actually doing the analysis yet.)
I’m not the most conventional mainstream thinker about random effects (see https://dynamicecology.wordpress.com/2015/11/04/is-it-a-fixed-or-random-effect/). But I think in some cases dimensions would be random effects (e.g. site or laboratory) but in other cases they could be fixed effects (e.g. species might often be a fixed effect but could easily be random too).
Comment 2 of 3.
I suggest adding two other descriptions of the instance-row, variable-column format:
1. There shall be a column that serves as a unique identifier.
2. Each row shall be fully populated: no blanks, no ambiguous coding like NA, 999, etc. Provide verbose descriptions (possibly using codes and footnotes) of each missing datum (e.g., respondent refused to answer, specimen not available, test failed, ran out of time, and so forth.)
I think those are good additions that maximize the benefit of a row-column format.
Comment 3 of 3.
Given the typical setup on our computers where MS Excel thinks .csv files are its own, it seems prudent to me to save delimited files (whether using commas or tabs as separators) with the .txt extension. If you start from Excel, save the file in .csv format, but then change the extension to .txt. That way, when you open the file again in Excel, you invoke the Text Import Wizard and have the opportunity to specify that a column is text (e.g., to preserve leading zeros in zip codes), date (with the opportunity to choose MDY versus DMY).
That makes sense to me.
Pingback: Digitando e conferindo dados – Mais Um Blog de Ecologia e Estatística
As a scientific researcher who has also constructed commercial and Governmental relational databases I strongly recommend that you make sure you understand relational database design as it is necessary to ensure data integrity. If you make the effort to understand the scientific basis of the relational data model you will understand that:
denormalization does not give better performance than a normalized database;
denormalization introduces data redundancy which means you lose single point of data maintenance and therefore on data updates your data can become inconsistent.
See
http://www.dbdebunk.com/2014/02/the-costly-denormalization-for.html
Yes, keep your raw data.
If you need data redundancy then achieve that with data backups.
Understanding the relational data model takes considerable time and effort, but if you really want to preserve your precious hard-won data from corruption and inconsistencies it is sensible to pay close attention to integrity of the bedrock of your research – your DATA! If you are currently accessing your data from Excel spreadsheets or .csv files or some other non-relational format then do not take my word for it, you will sooner or later SERIOUSLY regret it. Data structure is a non-trivial issue and cannot be ignored. It really concerns me that many people now put their precious data up in the ‘Cloud’ when there is absolutely no visibility as the reliability, backup and integrity of processes preserving the data.
Remember the scientific approach requires us to check every step of the argument.
There are plenty of good tutorials online and anyone who deals in data should at least learn to appreciate Codd’s rules.
As a scientific researcher who previously worked in the database industry for 9 years working on database internals and holding a patent in the area, I suggest you stop espousing dogma and actually slow down and read what I said.
If you read a little more closely, you will find that I am espousing denormalization in cases focused on analysis of data, not on so-called transactional systems that are focused on capturing new data with low fail rates. As you said it is basically a performance/integrity trade-off, but depending on the goals different positions along that trade-off axis are rational. What I’m proposing is not odd or contrary to practice at all. Datawarehousing regularly denormalizes. Its just a case of the right tool for the right job. There is no reason to assume databases designed for different purposes should be structured the same.
Now as for praising Codd’s rules, are you aware that he wrote 12 rules for OLAP (basically datawarehousing) that are quite different than his 12 rules for relational databases: http://olap.com/learn-bi-olap/codds-paper/ ?
I do agree with you about the excessive faith people have in the Cloud.
Thanks for these excellent, straightforward reference.
I’m trying to find a similar reference to share with my team that lays out as clearly as you have here for the data management/wrangling steps, a general workflow and best practices for the step required prior to these steps—i.e. data cleaning.
I know that it’s a highly variable task, but from my own experience there’s certain fundamental steps and considerations (e.g. make sure variables are stored in the proper format (num vs char), check that data are within expected ranges, check “if A then B” relationships, look for outliers, etc. etc. …)
There are piles of references on how to use various tools to perform the various required tasks (e.g. how to use dplyr to rearrange data into a tidy format, or assertr to test assumptions), but I haven’t found a solid, succinct reference on how to figure out what specific tasks should be performed to ensure that data is not just tidy, but also accurate/correct.
Anyone have any suggestions?
Hi Jonathan. Thanks for the positive feedback. I don’t know of such a resource either. I have had thoughts of writing such a post in the back of my mind for a while now. Maybe this will prompt me to do it. Or if you have interests in collaborating or writing such a post email me.
Pingback: Como organizar uma planilha de dados – Sobrevivendo na Ciência
Pingback: Como organizar uma planilha de dados | José Felipe de Almeida
Pingback: Organizando dados: de dados brutos a dados para análise, em R – Mais Um Blog de Ecologia e Estatística
Thank you for this post! I work in device analysis for a large scale semiconductor fabrication company, and everything you have written here is applicable for our parametrics as well! Would it be ok for me to share this post with some of our new hires (including your name and the link to this post)?